Chapter 2 Prepare and Describe the Data
This chapter prepares the data set and does some univariate and multivariate description of its characteristics prior to the CFA implementation in later chapters. Both numeric and graphical description and inference about distribution shape are quickly available with R functions from the psych and MVN packages.
2.1 The Data Set
The 175 case data set (no missing observations) is loaded from a .csv file. The .csv file was exported from the SPSS system file that is available from the website for the Tabachnick textbook (Tabachnick et al., 2019) It has eleven subscales from the WISC:
- info (Information)
- comp (Comprehension)
- arith (Arithmetic)
- simil (Similarities)
- vocab (Vocabulary)
- digit (Digit Span)
- pictcomp (Picture Completion)
- parang (Picture Arrangement)
- block (Block Design)
- object(Object Assembly)
- coding (Coding - not sure if it is A or B, or a combination)
The user may recognize these scales as commonly discussed subtests of the WISC. The first 6 variables comprise the set of manifest variables for the latent factor known as Verbal. The last five are associated with Performance.
The original data file also contains an ID variable that is dropped for the working object created as wisc2 here.
# import the primary data file
wisc1 <- read.csv("wisc1.csv")
knitr::kable(head(wisc1),booktabs=TRUE,format="markdown")| ID | info | comp | arith | simil | vocab | digit | pictcomp | parang | block | object | coding |
|---|---|---|---|---|---|---|---|---|---|---|---|
| 3 | 8 | 7 | 13 | 9 | 12 | 9 | 6 | 11 | 12 | 7 | 9 |
| 4 | 9 | 6 | 8 | 7 | 11 | 12 | 6 | 8 | 7 | 12 | 14 |
| 5 | 13 | 18 | 11 | 16 | 15 | 6 | 18 | 8 | 11 | 12 | 9 |
| 6 | 8 | 11 | 6 | 12 | 9 | 7 | 13 | 4 | 7 | 12 | 11 |
| 7 | 10 | 3 | 8 | 9 | 12 | 9 | 7 | 7 | 11 | 4 | 10 |
| 8 | 11 | 7 | 15 | 12 | 10 | 12 | 6 | 12 | 10 | 5 | 10 |
A note about tables in this document: Many of the tables generated by the various R functions in this document are reformatted so that they do not appear as the plain text that is typically output into the R console. The kable function in the knitr package permits formatting that is well-rendered with rmarkdown and bookdown document production. kable is used frequently.
2.2 Numeric and Graphical Description of the Data
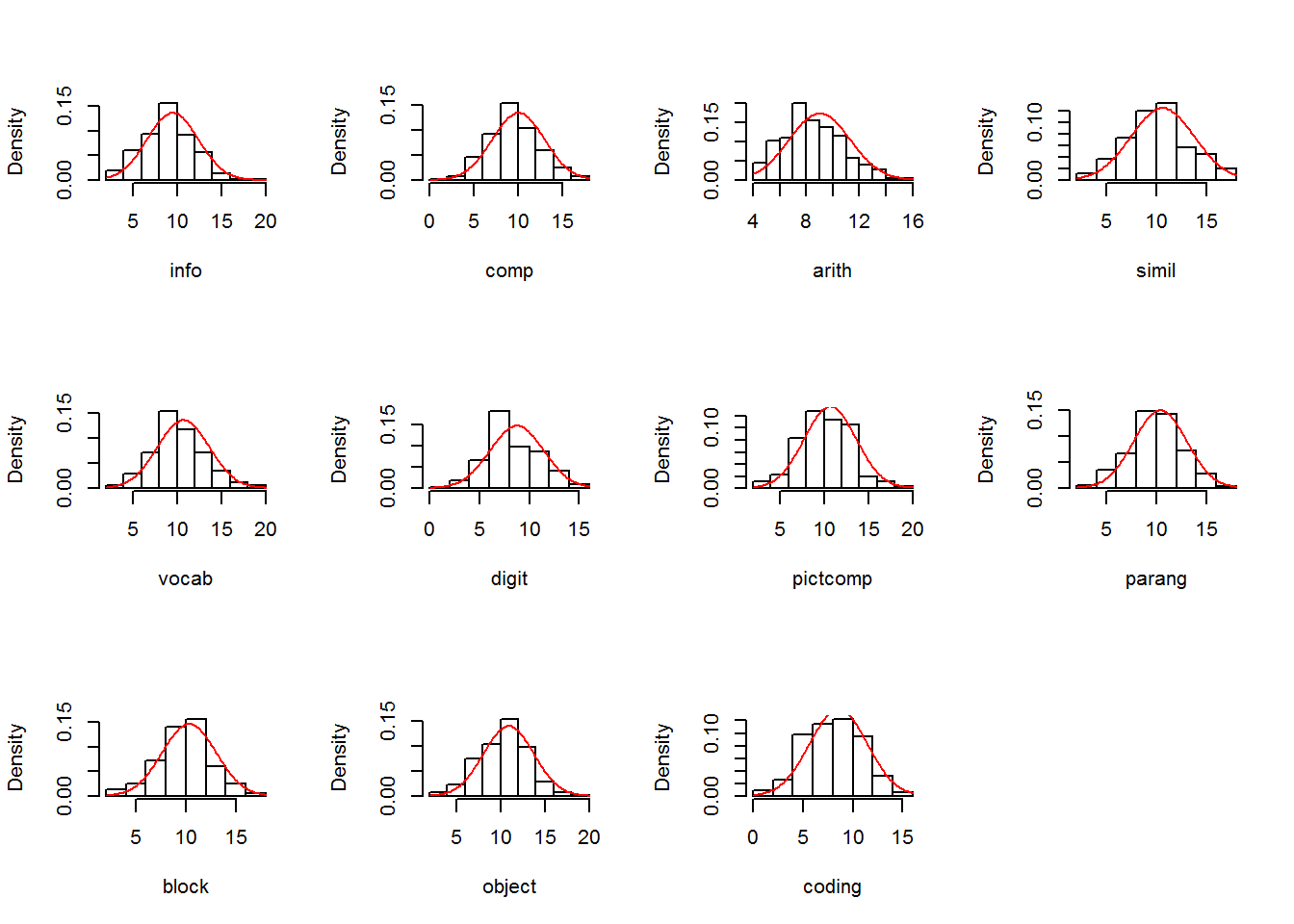
We can explore univariate characteristics of the data with summaries, plots, and evaluation of normality characteristics
2.2.1 Univariate descriptive statistics from the psych package.
| vars | n | mean | sd | median | min | max | range | skew | kurtosis | se | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| info | 1 | 175 | 9.497143 | 2.912269 | 10 | 3 | 19 | 16 | 0.0848560 | -0.0141461 | 0.2201469 |
| comp | 2 | 175 | 10.000000 | 2.965317 | 10 | 0 | 18 | 18 | 0.0869549 | 0.4102585 | 0.2241569 |
| arith | 3 | 175 | 9.000000 | 2.306911 | 9 | 4 | 16 | 12 | 0.3977676 | -0.1156374 | 0.1743861 |
| simil | 4 | 175 | 10.611429 | 3.183630 | 11 | 2 | 18 | 16 | 0.0227180 | -0.1686029 | 0.2406598 |
| vocab | 5 | 175 | 10.702857 | 2.932721 | 10 | 2 | 19 | 17 | 0.2716552 | 0.3699874 | 0.2216929 |
| digit | 6 | 175 | 8.731429 | 2.704166 | 8 | 0 | 16 | 16 | 0.2709366 | 0.1456805 | 0.2044157 |
| pictcomp | 7 | 175 | 10.680000 | 2.934221 | 11 | 2 | 19 | 17 | -0.0727446 | 0.3737787 | 0.2218063 |
| parang | 8 | 175 | 10.371429 | 2.659679 | 10 | 2 | 17 | 15 | -0.2025460 | 0.0097769 | 0.2010528 |
| block | 9 | 175 | 10.314286 | 2.709831 | 10 | 2 | 18 | 16 | -0.2235393 | 0.5932319 | 0.2048440 |
| object | 10 | 175 | 10.902857 | 2.843978 | 11 | 3 | 19 | 16 | -0.1250271 | 0.2241732 | 0.2149845 |
| coding | 11 | 175 | 8.548571 | 2.872118 | 9 | 0 | 15 | 15 | -0.0537486 | -0.4017079 | 0.2171117 |
2.2.2 Univariate Distribution Tests and Plots plus Evaluation of Multivariate Normality
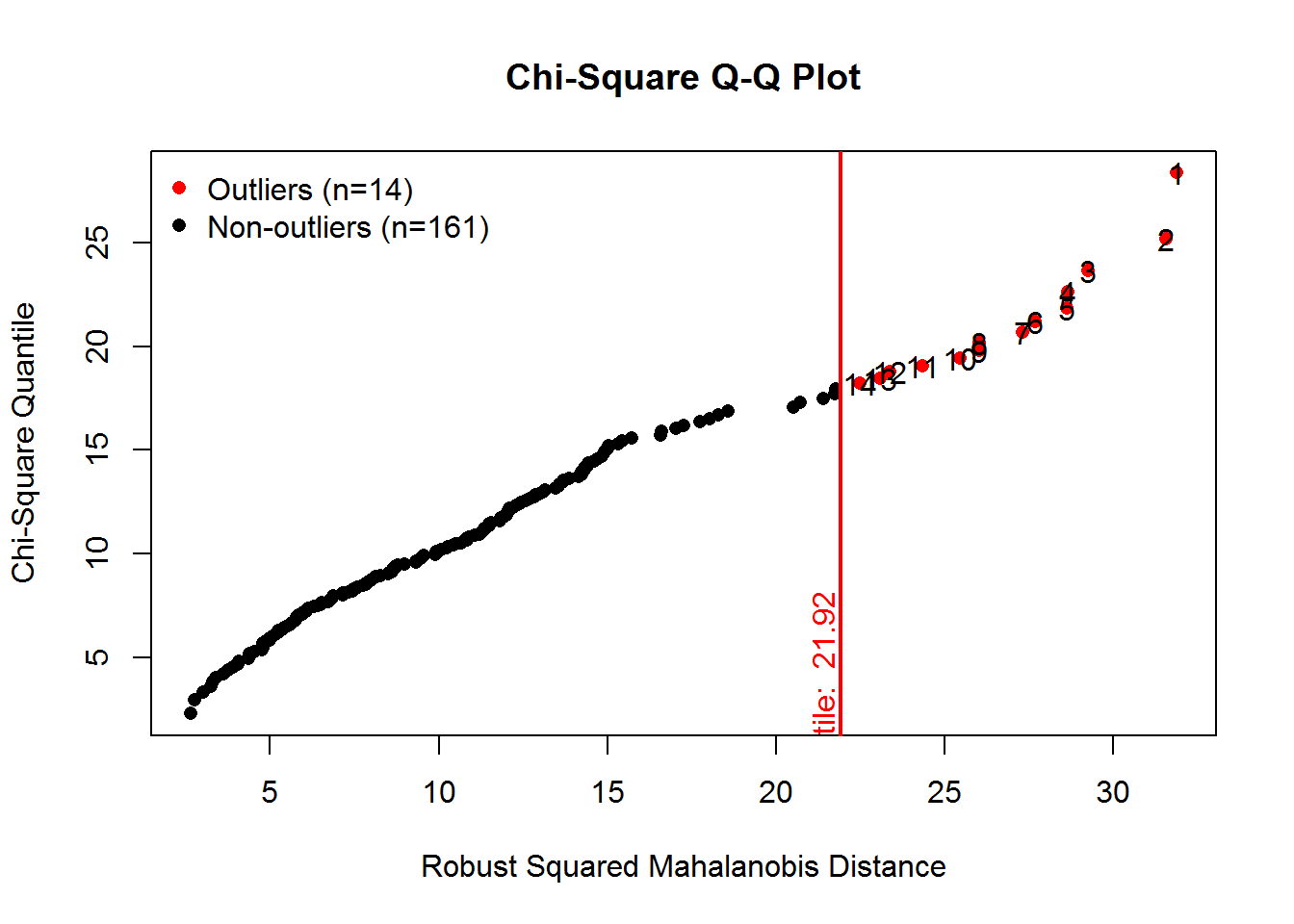
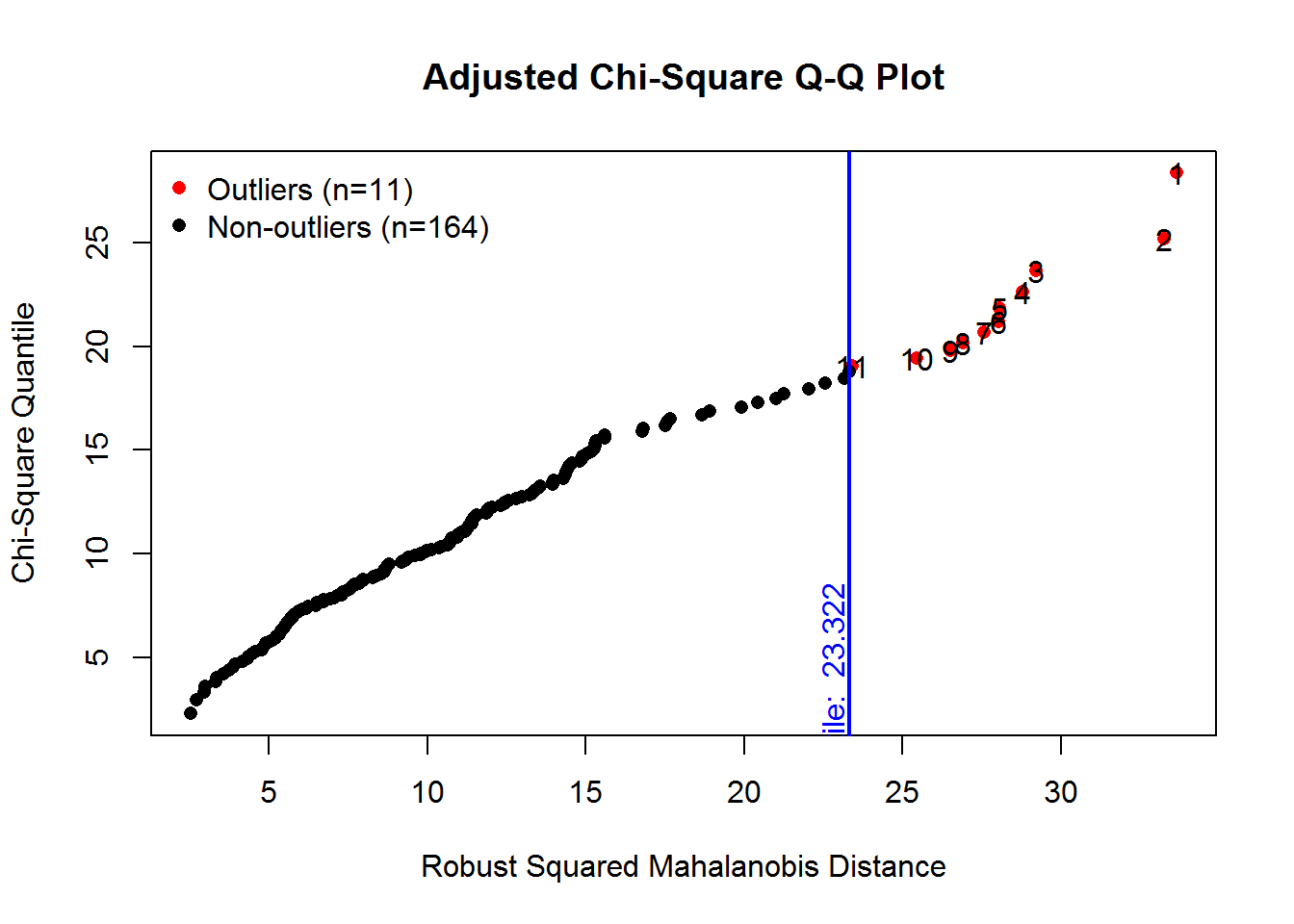
The MVN package provides univariate and multivariate normality tests. It is an efficient way to explore characteristics of a set of variables. Several options are available for testing both univariate and Multivariate normality. First, explicit calls for univariate and multivariate tests are made, and then an approach is shown that obtains all at once plus a useful set of plots.
x_vars <- wisc2
# use the mvn function for an extensive evaluation
# note that different kinds of tests can be specified with changes in the arguments
result <- mvn(data= x_vars, mvnTest="mardia", univariateTest="AD")
kable(result$univariateNormality, booktabs=TRUE, format="markdown")| Test | Variable | Statistic | p value | Normality |
|---|---|---|---|---|
| Anderson-Darling | info | 1.2049 | 0.0037 | NO |
| Anderson-Darling | comp | 1.3546 | 0.0016 | NO |
| Anderson-Darling | arith | 1.8656 | 1e-04 | NO |
| Anderson-Darling | simil | 0.9336 | 0.0175 | NO |
| Anderson-Darling | vocab | 1.4992 | 7e-04 | NO |
| Anderson-Darling | digit | 2.5087 | <0.001 | NO |
| Anderson-Darling | pictcomp | 1.1472 | 0.0052 | NO |
| Anderson-Darling | parang | 1.3391 | 0.0017 | NO |
| Anderson-Darling | block | 1.5225 | 6e-04 | NO |
| Anderson-Darling | object | 1.1593 | 0.0049 | NO |
| Anderson-Darling | coding | 1.2217 | 0.0034 | NO |
| Test | Statistic | p value | Result |
|---|---|---|---|
| Mardia Skewness | 348.496907430775 | 0.0067313733585278 | NO |
| Mardia Kurtosis | 2.30789388829536 | 0.0210050390817529 | NO |
| MVN | NA | NA | NO |
|
|
|
2.3 Bivariate Characteristics of the data set
We can quickly explore numerical and graphical summaries of the eleven variables.
2.3.1 SPLOM
Among the many scatterplot matrix capabilies in R, John Fox’ scatterplot.matrix function in his car package has probably been seen by most students.
scatterplotMatrix(wisc2,cex=.2,
smooth=list(col.smooth="red", spread=F, lwd.smooth=.3),
col="skyblue1",
regLine=list(lwd=.3,col="black"))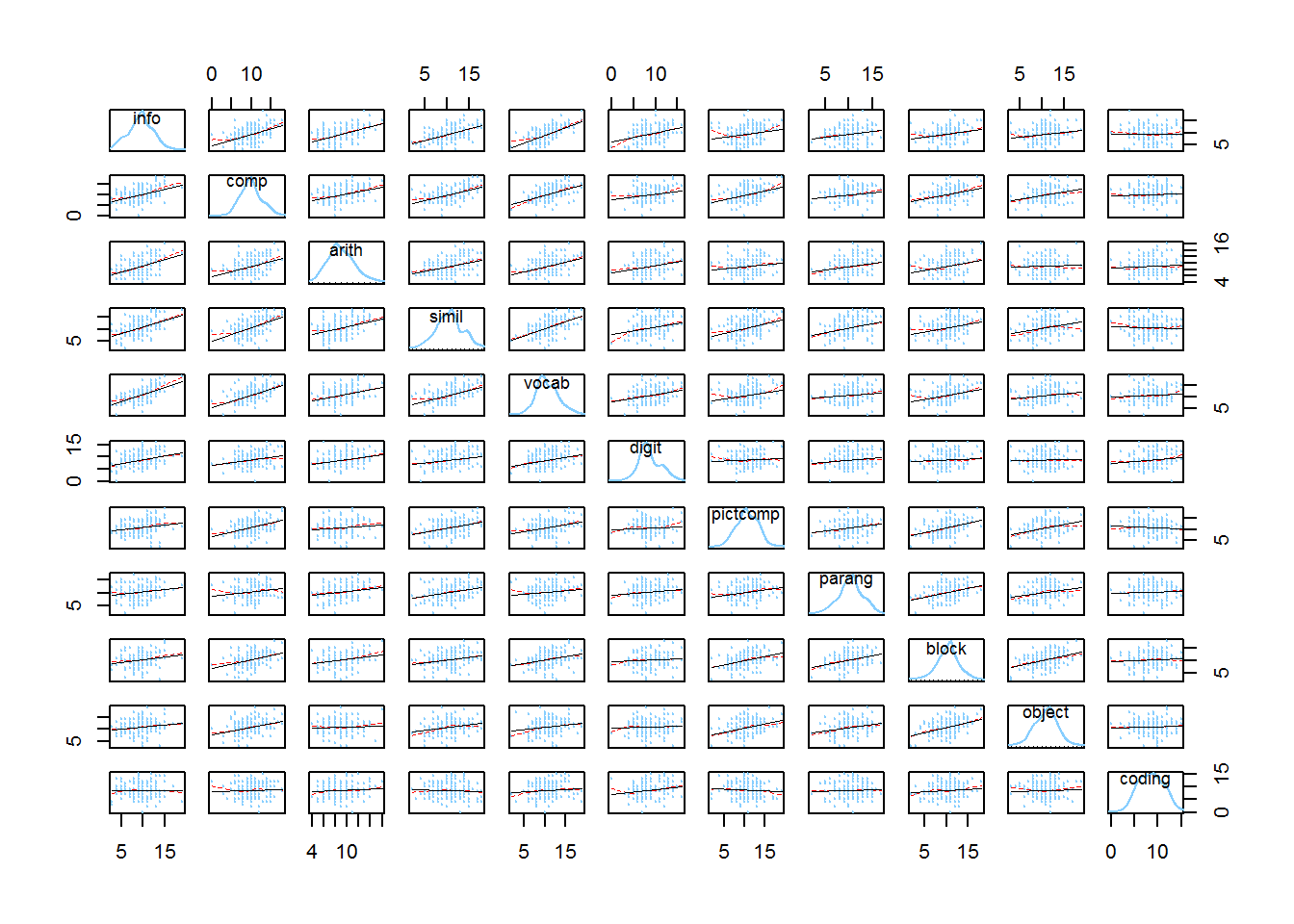
Even with some control over colors and sizes of points/lines, this SPLOM probably has too many variables to be effective - each plot is very small. Nonetheless, the sense of fairly linear relationships among all pairs is somewhat apparent, as is the relative univariate normality of each of the eleven.
Note that the image can be enlarged if the reader is using a pdf version of this document simply by using the increase/decrease size capability of pdf readers. If the user is reading an html version of this document, then try to do a right mouse click on the image and “view image” (in Windows). Then the image can be increased in size in a browser.
2.4 Covariances and Zero Order Correlations
The covariance matrix is the basic input for the CFA algorithms outlined in later chapters.
| info | comp | arith | simil | vocab | digit | pictcomp | parang | block | object | coding | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| info | 8.481 | 4.034 | 3.322 | 4.758 | 5.338 | 2.720 | 1.965 | 1.561 | 1.808 | 1.531 | 0.059 |
| comp | 4.034 | 8.793 | 2.684 | 4.816 | 4.621 | 1.891 | 3.540 | 1.471 | 2.966 | 2.718 | 0.517 |
| arith | 3.322 | 2.684 | 5.322 | 2.713 | 2.621 | 1.678 | 1.052 | 1.391 | 1.701 | 0.282 | 0.598 |
| simil | 4.758 | 4.816 | 2.713 | 10.136 | 5.022 | 2.234 | 3.450 | 2.524 | 2.255 | 2.433 | -0.372 |
| vocab | 5.338 | 4.621 | 2.621 | 5.022 | 8.601 | 2.334 | 2.456 | 1.031 | 2.364 | 1.546 | 0.842 |
| digit | 2.720 | 1.891 | 1.678 | 2.234 | 2.334 | 7.313 | 0.597 | 1.066 | 0.533 | 0.267 | 1.344 |
| pictcomp | 1.965 | 3.540 | 1.052 | 3.450 | 2.456 | 0.597 | 8.610 | 1.941 | 3.038 | 3.032 | -0.605 |
| parang | 1.561 | 1.471 | 1.391 | 2.524 | 1.031 | 1.066 | 1.941 | 7.074 | 2.532 | 1.916 | 0.289 |
| block | 1.808 | 2.966 | 1.701 | 2.255 | 2.364 | 0.533 | 3.038 | 2.532 | 7.343 | 3.077 | 0.832 |
| object | 1.531 | 2.718 | 0.282 | 2.433 | 1.546 | 0.267 | 3.032 | 1.916 | 3.077 | 8.088 | 0.433 |
| coding | 0.059 | 0.517 | 0.598 | -0.372 | 0.842 | 1.344 | -0.605 | 0.289 | 0.832 | 0.433 | 8.249 |
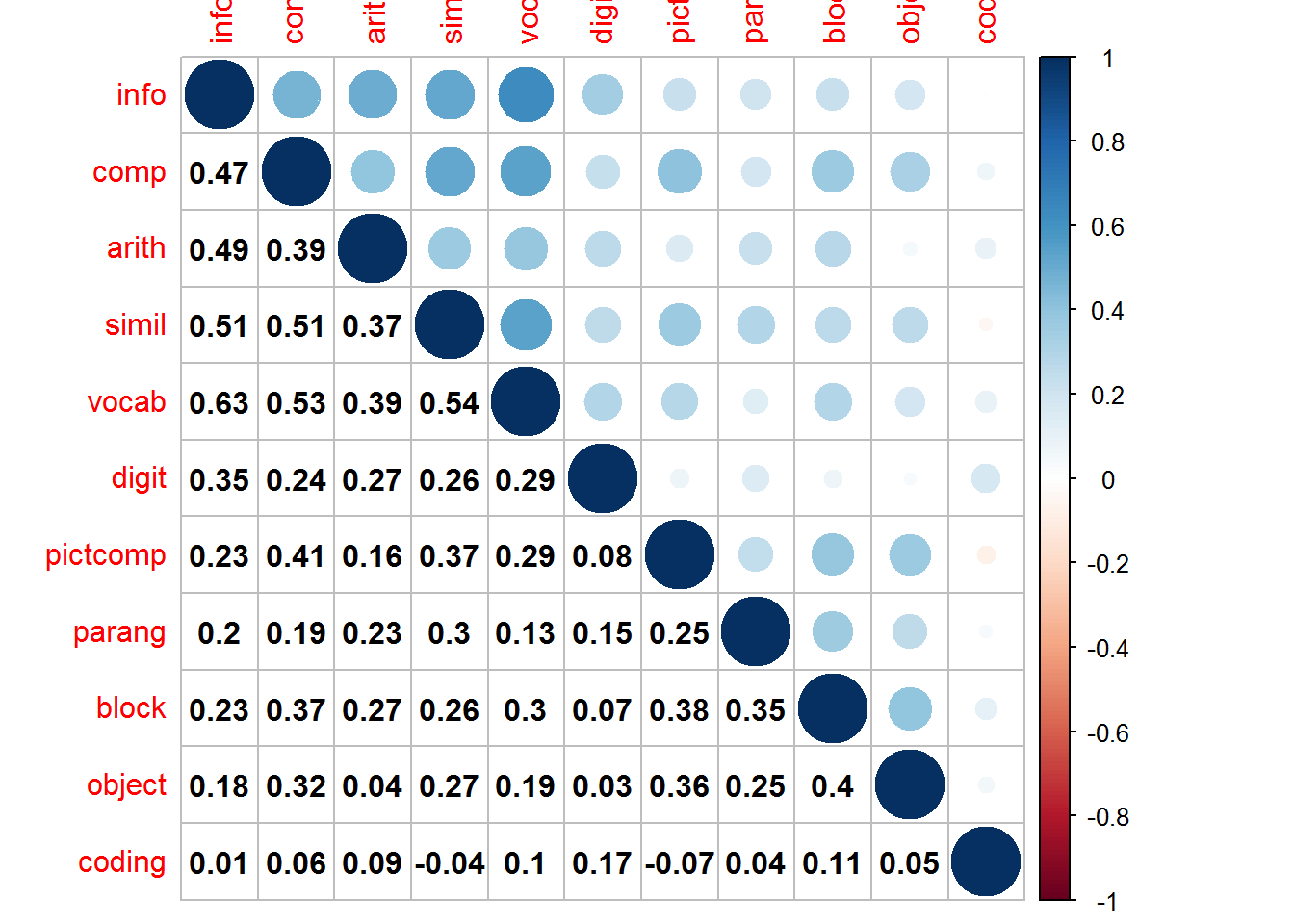
We can use the Corrplot package to produce a useful combination of a schematic and correlation matrix.
mat1 <- cor(wisc2)
corrplot(mat1,type="upper",tl.pos="tp")
corrplot(mat1,add=T,type="lower", method="number",
col="black", diag=FALSE,tl.pos="n", cl.pos="n")