Chapter 3 Using the lavaan package for CFA
One of the primary tools for SEM in R is the lavaan package. It permits path specification with a simple syntax.
3.1 Implement the CFA, First Model
Using the lavaan package, we can implemnt directly the CFA with only a few steps. Since this document contains three different packages’ approach to CFA, the packages used for each will be loaded at that point, so as to not have confusion over common function names.
3.1.1 Define and fit the first model
The latent variables and their paths to the manifest variables are initially defined as simple textual specifications. The =~ symbol is the key to defining paths. We have two latent variables and no manifest variable has duplicate paths from both latents. This is a so-called “simple structure.”
Note that this text string uses variable names that match what is in the wisc2 data set.
wisc.model1 <- "verbal =~ info + comp + arith + simil + vocab + digit
performance =~ pictcomp + parang + block + object + coding"Fit the model and obtain the basic summary. Note that in this default approach, the latent factors are permitted to covary and the model estimates this covariance.
One R syntax note….. the format here to call the cfa function (lavaan::cfa(.....)) is employed to ensure no ambiguity that the correct cfa function is the one from the lavaan package. This precludes confusion when multiple packages contain functions with the same name as is the case with both lavaan and sem which is also used in this document. Even though sem is loaded later in this document, if there is a chance that it may simultaneously exist in the R environment with lavaan then the approach here is safer.
fit1 <- lavaan::cfa(wisc.model1, data=wisc2,std.lv=TRUE)
summary(fit1, fit.measures=T,standardized=T)## lavaan 0.6-19 ended normally after 24 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 23
##
## Number of observations 175
##
## Model Test User Model:
##
## Test statistic 70.640
## Degrees of freedom 43
## P-value (Chi-square) 0.005
##
## Model Test Baseline Model:
##
## Test statistic 519.204
## Degrees of freedom 55
## P-value 0.000
##
## User Model versus Baseline Model:
##
## Comparative Fit Index (CFI) 0.940
## Tucker-Lewis Index (TLI) 0.924
##
## Loglikelihood and Information Criteria:
##
## Loglikelihood user model (H0) -4491.822
## Loglikelihood unrestricted model (H1) -4456.502
##
## Akaike (AIC) 9029.643
## Bayesian (BIC) 9102.433
## Sample-size adjusted Bayesian (SABIC) 9029.600
##
## Root Mean Square Error of Approximation:
##
## RMSEA 0.061
## 90 Percent confidence interval - lower 0.033
## 90 Percent confidence interval - upper 0.085
## P-value H_0: RMSEA <= 0.050 0.233
## P-value H_0: RMSEA >= 0.080 0.103
##
## Standardized Root Mean Square Residual:
##
## SRMR 0.059
##
## Parameter Estimates:
##
## Standard errors Standard
## Information Expected
## Information saturated (h1) model Structured
##
## Latent Variables:
## Estimate Std.Err z-value P(>|z|) Std.lv Std.all
## verbal =~
## info 2.206 0.200 11.029 0.000 2.206 0.760
## comp 2.042 0.210 9.709 0.000 2.042 0.691
## arith 1.300 0.172 7.555 0.000 1.300 0.565
## simil 2.232 0.225 9.940 0.000 2.232 0.703
## vocab 2.250 0.200 11.225 0.000 2.250 0.770
## digit 1.053 0.212 4.967 0.000 1.053 0.390
## performance =~
## pictcomp 1.742 0.242 7.187 0.000 1.742 0.595
## parang 1.253 0.224 5.582 0.000 1.253 0.473
## block 1.846 0.222 8.311 0.000 1.846 0.683
## object 1.605 0.236 6.800 0.000 1.605 0.566
## coding 0.207 0.254 0.814 0.416 0.207 0.072
##
## Covariances:
## Estimate Std.Err z-value P(>|z|) Std.lv Std.all
## verbal ~~
## performance 0.589 0.075 7.814 0.000 0.589 0.589
##
## Variances:
## Estimate Std.Err z-value P(>|z|) Std.lv Std.all
## .info 3.566 0.507 7.034 0.000 3.566 0.423
## .comp 4.572 0.585 7.815 0.000 4.572 0.523
## .arith 3.602 0.420 8.571 0.000 3.602 0.681
## .simil 5.096 0.662 7.702 0.000 5.096 0.506
## .vocab 3.487 0.506 6.886 0.000 3.487 0.408
## .digit 6.162 0.680 9.056 0.000 6.162 0.848
## .pictcomp 5.526 0.757 7.296 0.000 5.526 0.646
## .parang 5.463 0.658 8.298 0.000 5.463 0.777
## .block 3.894 0.640 6.083 0.000 3.894 0.533
## .object 5.467 0.719 7.600 0.000 5.467 0.680
## .coding 8.159 0.874 9.335 0.000 8.159 0.995
## verbal 1.000 1.000 1.000
## performance 1.000 1.000 1.0003.1.2 Obtain coefficients
It is possible to obtain the output from the above summary function that does not contain the parameter coefficients part of the table. If that had been the implementation, then the coefficients can be obtained simply, but it is better to obtain the full table of parameter estimates and errors, etc.
3.1.3 Complete parameter listing
Instead of directly printing the parameter table, I prefer to reformat it with kable when using R Markdown. Without using kable, the code would be the following:
Format the table and clean it up using kable.
parameterEstimates(fit1, standardized=TRUE) %>%
filter(op == "=~") %>%
select('Latent Factor'=lhs, Indicator=rhs, B=est, SE=se, Z=z, 'p-value'=pvalue, Beta=std.all) %>%
knitr::kable(digits = 3, booktabs=TRUE, format="markdown", caption="Factor Loadings")| Latent Factor | Indicator | B | SE | Z | p-value | Beta |
|---|---|---|---|---|---|---|
| verbal | info | 2.206 | 0.200 | 11.029 | 0.000 | 0.760 |
| verbal | comp | 2.042 | 0.210 | 9.709 | 0.000 | 0.691 |
| verbal | arith | 1.300 | 0.172 | 7.555 | 0.000 | 0.565 |
| verbal | simil | 2.232 | 0.225 | 9.940 | 0.000 | 0.703 |
| verbal | vocab | 2.250 | 0.200 | 11.225 | 0.000 | 0.770 |
| verbal | digit | 1.053 | 0.212 | 4.967 | 0.000 | 0.390 |
| performance | pictcomp | 1.742 | 0.242 | 7.187 | 0.000 | 0.595 |
| performance | parang | 1.253 | 0.224 | 5.582 | 0.000 | 0.473 |
| performance | block | 1.846 | 0.222 | 8.311 | 0.000 | 0.683 |
| performance | object | 1.605 | 0.236 | 6.800 | 0.000 | 0.566 |
| performance | coding | 0.207 | 0.254 | 0.814 | 0.416 | 0.072 |
3.1.4 Residuals correlation matrix
Residuals can be examined.
cor_table <- residuals(fit1, type = "cor")$cov
#cor_table[upper.tri(cor_table)] <- # erase the upper triangle
#diag(cor_table) <- NA # erase the diagonal 0's
knitr::kable(cor_table, digits=3,format="markdown", booktabs=TRUE) # makes a nice table and rounds everyhing to 2 digits| info | comp | arith | simil | vocab | digit | pictcomp | parang | block | object | coding | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| info | 0.000 | -0.058 | 0.065 | -0.021 | 0.040 | 0.049 | -0.036 | -0.010 | -0.076 | -0.068 | -0.025 |
| comp | -0.058 | 0.000 | 0.002 | 0.025 | 0.000 | -0.034 | 0.165 | -0.006 | 0.091 | 0.092 | 0.031 |
| arith | 0.065 | 0.002 | 0.000 | -0.028 | -0.047 | 0.048 | -0.043 | 0.069 | 0.045 | -0.145 | 0.066 |
| simil | -0.021 | 0.025 | -0.028 | 0.000 | -0.003 | -0.015 | 0.123 | 0.102 | -0.021 | 0.034 | -0.071 |
| vocab | 0.040 | 0.000 | -0.047 | -0.003 | 0.000 | -0.006 | 0.016 | -0.082 | -0.012 | -0.071 | 0.067 |
| digit | 0.049 | -0.034 | 0.048 | -0.015 | -0.006 | 0.000 | -0.062 | 0.040 | -0.084 | -0.095 | 0.156 |
| pictcomp | -0.036 | 0.165 | -0.043 | 0.123 | 0.016 | -0.062 | 0.000 | -0.033 | -0.025 | 0.026 | -0.115 |
| parang | -0.010 | -0.006 | 0.069 | 0.102 | -0.082 | 0.040 | -0.033 | 0.000 | 0.028 | -0.014 | 0.004 |
| block | -0.076 | 0.091 | 0.045 | -0.021 | -0.012 | -0.084 | -0.025 | 0.028 | 0.000 | 0.013 | 0.058 |
| object | -0.068 | 0.092 | -0.145 | 0.034 | -0.071 | -0.095 | 0.026 | -0.014 | 0.013 | 0.000 | 0.012 |
| coding | -0.025 | 0.031 | 0.066 | -0.071 | 0.067 | 0.156 | -0.115 | 0.004 | 0.058 | 0.012 | 0.000 |
3.1.5 Plot the residuals.
norm plot
# extract the residuals from the fit1 model
# get rid of the duplicates and diagonal values
# create a vector for a
res1 <- residuals(fit1, type = "cor")$cov
res1[upper.tri(res1,diag=T)] <- NA
v1 <- as.vector(res1)
v2 <- v1[!is.na(v1)]
qqPlot(v2,id=F)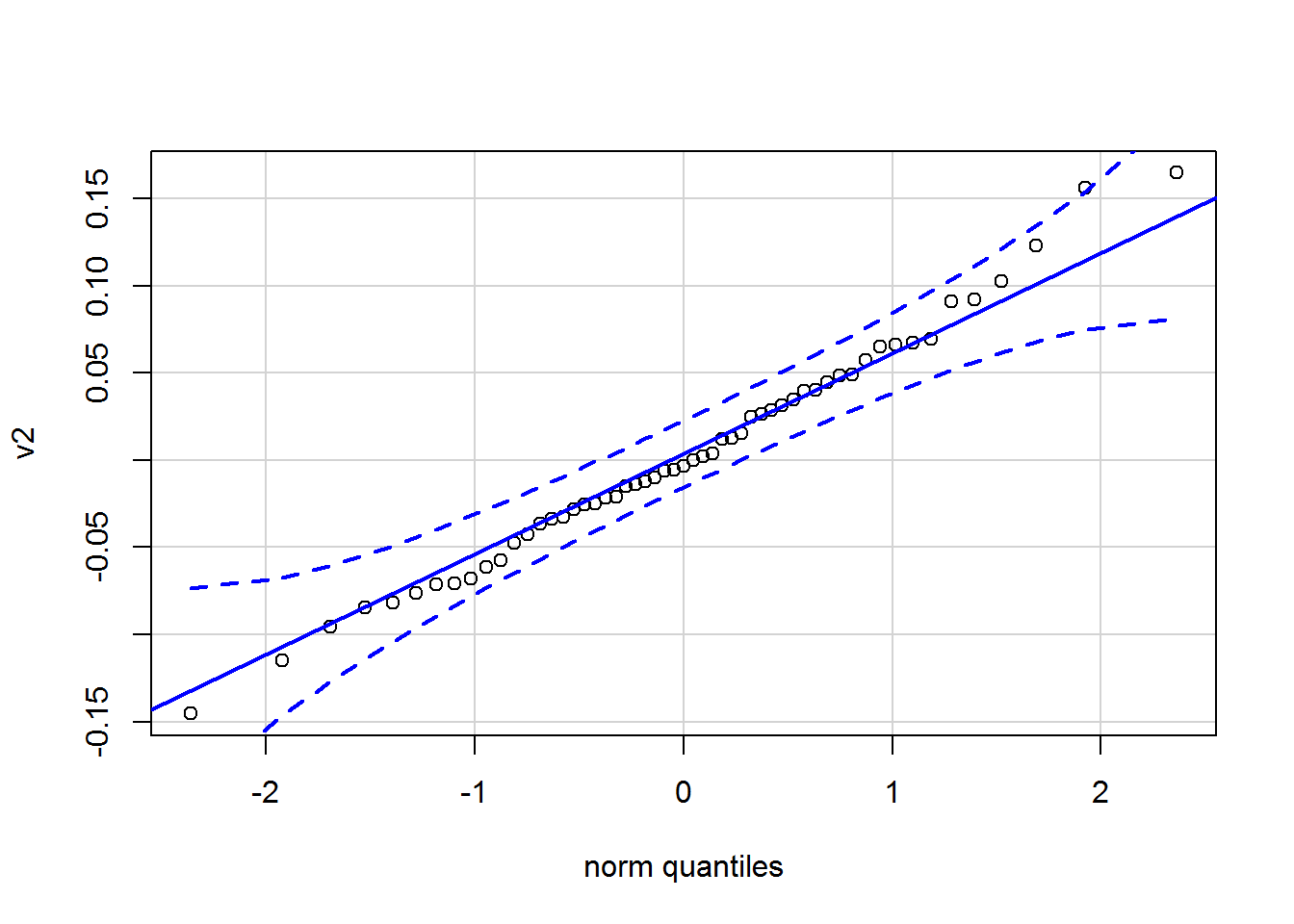
3.1.6 Modification Indices
Modification indices provide……
| lhs | op | rhs | mi | epc | sepc.lv | sepc.all | sepc.nox | |
|---|---|---|---|---|---|---|---|---|
| 32 | performance | =~ | comp | 9.823223 | 0.9372111 | 0.9372111 | 0.3169647 | 0.3169647 |
| 62 | arith | ~~ | object | 6.362360 | -0.9445341 | -0.9445341 | -0.2128471 | -0.2128471 |
| 37 | info | ~~ | comp | 5.221423 | -0.9847261 | -0.9847261 | -0.2438796 | -0.2438796 |
| 81 | digit | ~~ | coding | 4.926741 | 1.2072274 | 1.2072274 | 0.1702526 | 0.1702526 |
| 51 | comp | ~~ | pictcomp | 4.685434 | 0.9693124 | 0.9693124 | 0.1928391 | 0.1928391 |
| 85 | pictcomp | ~~ | coding | 4.574821 | -1.2249127 | -1.2249127 | -0.1824249 | -0.1824249 |
| 31 | performance | =~ | info | 4.476617 | -0.5955241 | -0.5955241 | -0.2050748 | -0.2050748 |
| 40 | info | ~~ | vocab | 4.402900 | 0.9124221 | 0.9124221 | 0.2587532 | 0.2587532 |
| 73 | vocab | ~~ | parang | 4.145852 | -0.7977279 | -0.7977279 | -0.1827712 | -0.1827712 |
| 38 | info | ~~ | arith | 4.133326 | 0.6970216 | 0.6970216 | 0.1944960 | 0.1944960 |
| 67 | simil | ~~ | parang | 3.373228 | 0.8304961 | 0.8304961 | 0.1574097 | 0.1574097 |
| 70 | simil | ~~ | coding | 3.271062 | -0.9560218 | -0.9560218 | -0.1482698 | -0.1482698 |
| 66 | simil | ~~ | pictcomp | 3.269521 | 0.8602498 | 0.8602498 | 0.1621163 | 0.1621163 |
3.1.7 Path Diagram for the bifactor Model 1
The semPlot package provides a capability of drawing path diagrams for cfa and other sem models. The semPaths function will take a cfa model object and draw the diagram, with several options available. The diagram produced here takes control over font/label sizes, display of residuals, and color of paths/coefficients. These and many more control options are available. There is a challenge in producing these path diagrams to have font sizes large enough for most humans to read. I’ve taken control of the font sizes for the “edges” with a cex argument. But this causes overlap in the values if the default layout is used. I found that “circle2” worked best here.
# Note that the base plot, including standardized path coefficients plots positive coefficients green
# and negative coefficients red. Red-green colorblindness issues anyone?
# I redrew it here to choose a blue and red. But all the coefficients in this example are
# positive,so they are shown with the skyblue.
# more challenging to use colors other than red and green. not in this doc
semPaths(fit1, residuals=F,sizeMan=7,"std",
posCol=c("skyblue4", "red"),
#edge.color="skyblue4",
edge.label.cex=1.2,layout="circle2")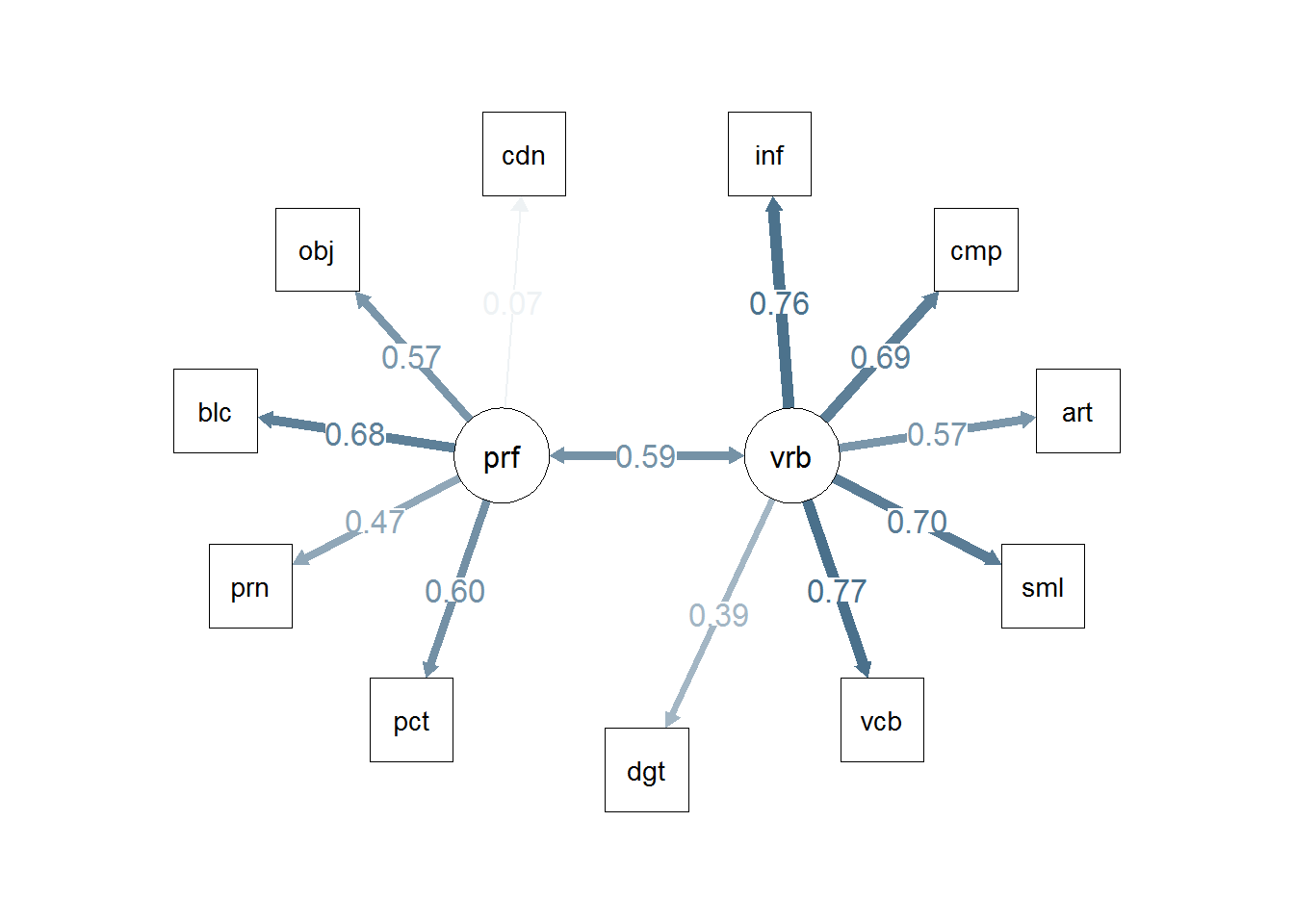
# or we could draw the paths in such a way to include the residuals:
#semPaths(fit1, sizeMan=7,"std",edge.color="skyblue4",edge.label.cex=1,layout="circle2")
# the base path diagram can be drawn much more simply:
#semPaths(fit1)
# or
semPaths(fit1,"std")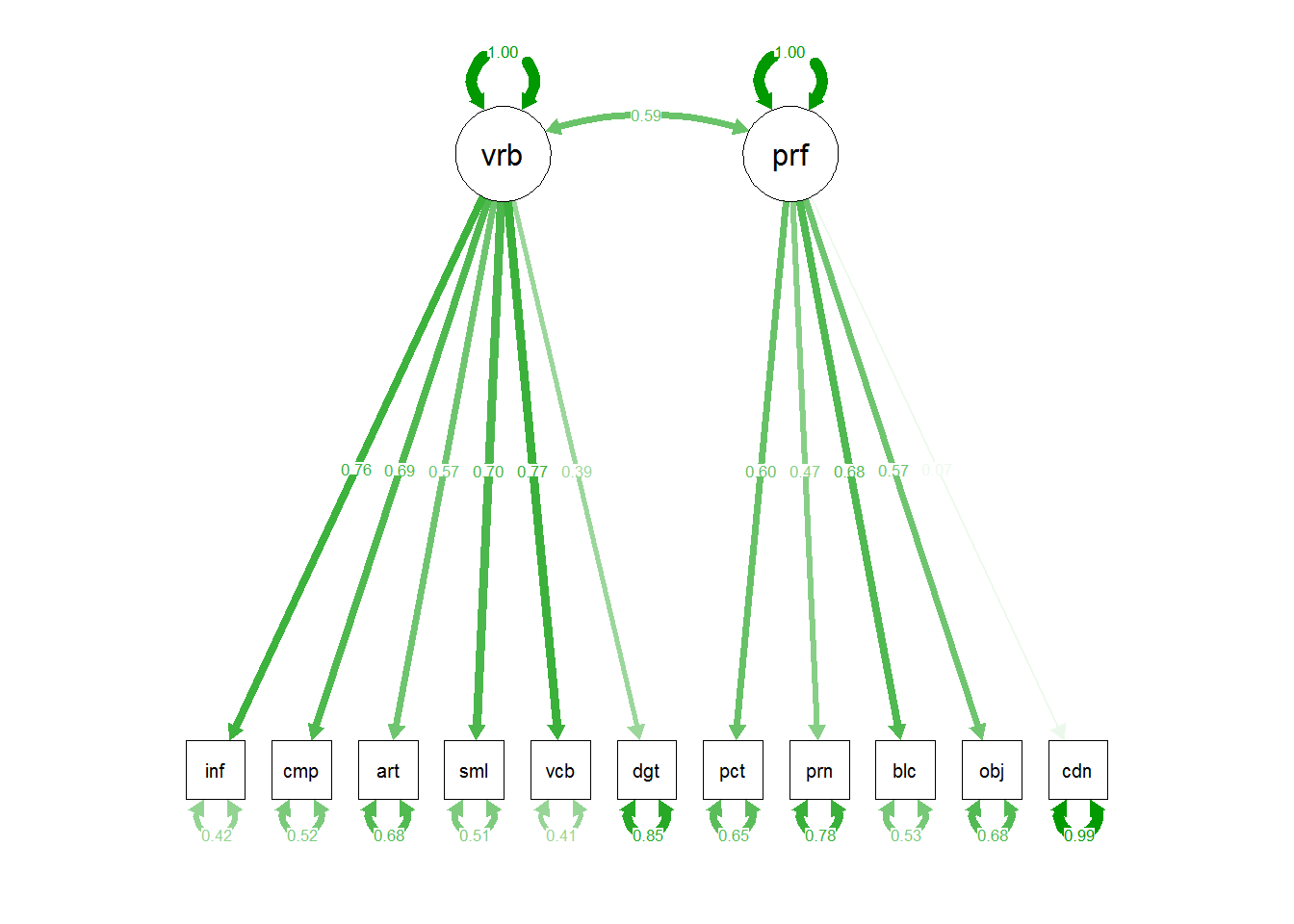
3.1.8 orthognal fit comparison
Compare to a one factor solution.
fit1orth <- lavaan::cfa(wisc.model1, data=wisc1,orthogonal=T)
kable(anova(fit1,fit1orth), booktabs=TRUE, format="markdown")| Df | AIC | BIC | Chisq | Chisq diff | RMSEA | Df diff | Pr(>Chisq) | |
|---|---|---|---|---|---|---|---|---|
| fit1 | 43 | 9029.643 | 9102.433 | 70.63957 | NA | NA | NA | NA |
| fit1orth | 44 | 9065.614 | 9135.239 | 108.60977 | 37.97019 | 0.4596284 | 1 | 0 |
3.2 Generate a second model and compare
Next, generate a second CFA model. Thre are theoretical reasons why paths from both latent factors to “comp” might be warranted. The “comp” variable also has the largest Modification index in the first model. This second modelimplements this one-path/parmaeter change.
3.2.1 Add a path (Perf to comp) and Fit the second CFA model
Define the addditional path in the model text string.
wisc.model2 <- 'verbal =~ info + comp + arith + simil + vocab + digit
performance =~ pictcomp + parang + block + object + coding + comp'Fit the model and obtain the basic summary
fit2 <- lavaan::cfa(wisc.model2, data=wisc1,std.lv=TRUE)
summary(fit2, fit.measures=T,standardized=T)## lavaan 0.6-19 ended normally after 26 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 24
##
## Number of observations 175
##
## Model Test User Model:
##
## Test statistic 60.642
## Degrees of freedom 42
## P-value (Chi-square) 0.031
##
## Model Test Baseline Model:
##
## Test statistic 519.204
## Degrees of freedom 55
## P-value 0.000
##
## User Model versus Baseline Model:
##
## Comparative Fit Index (CFI) 0.960
## Tucker-Lewis Index (TLI) 0.947
##
## Loglikelihood and Information Criteria:
##
## Loglikelihood user model (H0) -4486.823
## Loglikelihood unrestricted model (H1) -4456.502
##
## Akaike (AIC) 9021.645
## Bayesian (BIC) 9097.600
## Sample-size adjusted Bayesian (SABIC) 9021.600
##
## Root Mean Square Error of Approximation:
##
## RMSEA 0.050
## 90 Percent confidence interval - lower 0.016
## 90 Percent confidence interval - upper 0.077
## P-value H_0: RMSEA <= 0.050 0.465
## P-value H_0: RMSEA >= 0.080 0.031
##
## Standardized Root Mean Square Residual:
##
## SRMR 0.054
##
## Parameter Estimates:
##
## Standard errors Standard
## Information Expected
## Information saturated (h1) model Structured
##
## Latent Variables:
## Estimate Std.Err z-value P(>|z|) Std.lv Std.all
## verbal =~
## info 2.256 0.199 11.318 0.000 2.256 0.777
## comp 1.491 0.254 5.877 0.000 1.491 0.504
## arith 1.307 0.172 7.584 0.000 1.307 0.568
## simil 2.205 0.226 9.748 0.000 2.205 0.695
## vocab 2.273 0.201 11.329 0.000 2.273 0.777
## digit 1.075 0.212 5.068 0.000 1.075 0.399
## performance =~
## pictcomp 1.790 0.239 7.495 0.000 1.790 0.612
## parang 1.189 0.224 5.317 0.000 1.189 0.448
## block 1.823 0.219 8.334 0.000 1.823 0.675
## object 1.633 0.233 7.010 0.000 1.633 0.576
## coding 0.200 0.253 0.793 0.428 0.200 0.070
## comp 0.884 0.266 3.324 0.001 0.884 0.299
##
## Covariances:
## Estimate Std.Err z-value P(>|z|) Std.lv Std.all
## verbal ~~
## performance 0.533 0.081 6.594 0.000 0.533 0.533
##
## Variances:
## Estimate Std.Err z-value P(>|z|) Std.lv Std.all
## .info 3.343 0.502 6.665 0.000 3.343 0.396
## .comp 4.331 0.554 7.819 0.000 4.331 0.495
## .arith 3.584 0.420 8.533 0.000 3.584 0.677
## .simil 5.217 0.675 7.726 0.000 5.217 0.518
## .vocab 3.383 0.508 6.655 0.000 3.383 0.396
## .digit 6.116 0.677 9.032 0.000 6.116 0.841
## .pictcomp 5.358 0.741 7.231 0.000 5.358 0.626
## .parang 5.620 0.663 8.480 0.000 5.620 0.799
## .block 3.979 0.623 6.385 0.000 3.979 0.545
## .object 5.376 0.707 7.603 0.000 5.376 0.668
## .coding 8.162 0.874 9.337 0.000 8.162 0.995
## verbal 1.000 1.000 1.000
## performance 1.000 1.000 1.0003.2.2 Obtain coefficients
Coefficients can be obtained simply with the coef function, but it is better to do the full parameter listing in the next section
It is simple to obtain the full parameter list, but I prefer to use kable for tables when I can.
Reformat the table and clean it up by using kable.
parameterEstimates(fit2, standardized=TRUE) %>%
filter(op == "=~") %>%
select('Latent Factor'=lhs, Indicator=rhs, B=est, SE=se, Z=z, 'p-value'=pvalue, Beta=std.all) %>%
knitr::kable(digits = 3, format="markdown", booktabs=TRUE, caption="Factor Loadings")| Latent Factor | Indicator | B | SE | Z | p-value | Beta |
|---|---|---|---|---|---|---|
| verbal | info | 2.256 | 0.199 | 11.318 | 0.000 | 0.777 |
| verbal | comp | 1.491 | 0.254 | 5.877 | 0.000 | 0.504 |
| verbal | arith | 1.307 | 0.172 | 7.584 | 0.000 | 0.568 |
| verbal | simil | 2.205 | 0.226 | 9.748 | 0.000 | 0.695 |
| verbal | vocab | 2.273 | 0.201 | 11.329 | 0.000 | 0.777 |
| verbal | digit | 1.075 | 0.212 | 5.068 | 0.000 | 0.399 |
| performance | pictcomp | 1.790 | 0.239 | 7.495 | 0.000 | 0.612 |
| performance | parang | 1.189 | 0.224 | 5.317 | 0.000 | 0.448 |
| performance | block | 1.823 | 0.219 | 8.334 | 0.000 | 0.675 |
| performance | object | 1.633 | 0.233 | 7.010 | 0.000 | 0.576 |
| performance | coding | 0.200 | 0.253 | 0.793 | 0.428 | 0.070 |
| performance | comp | 0.884 | 0.266 | 3.324 | 0.001 | 0.299 |
3.2.3 Residuals correlation matrix
Residuals can be examined.
cor_table2 <- residuals(fit2, type = "cor")$cov
#cor_table[upper.tri(cor_table)] <- # erase the upper triangle
#diag(cor_table) <- NA # erase the diagonal 0's
knitr::kable(cor_table2, digits=3,format="markdown",booktabs=TRUE) # makes a nice table and rounds everyhing to 2 digits| info | comp | arith | simil | vocab | digit | pictcomp | parang | block | object | coding | |
|---|---|---|---|---|---|---|---|---|---|---|---|
| info | 0.000 | -0.049 | 0.053 | -0.026 | 0.021 | 0.036 | -0.023 | 0.016 | -0.050 | -0.054 | -0.022 |
| comp | -0.049 | 0.000 | 0.015 | 0.049 | 0.015 | -0.029 | 0.059 | -0.068 | -0.014 | -0.005 | 0.021 |
| arith | 0.053 | 0.015 | 0.000 | -0.025 | -0.054 | 0.043 | -0.030 | 0.091 | 0.068 | -0.131 | 0.069 |
| simil | -0.026 | 0.049 | -0.025 | 0.000 | -0.002 | -0.017 | 0.143 | 0.132 | 0.012 | 0.056 | -0.067 |
| vocab | 0.021 | 0.015 | -0.054 | -0.002 | 0.000 | -0.016 | 0.032 | -0.054 | 0.018 | -0.053 | 0.071 |
| digit | 0.036 | -0.029 | 0.043 | -0.017 | -0.016 | 0.000 | -0.055 | 0.053 | -0.071 | -0.088 | 0.158 |
| pictcomp | -0.023 | 0.059 | -0.030 | 0.143 | 0.032 | -0.055 | 0.000 | -0.025 | -0.031 | 0.011 | -0.115 |
| parang | 0.016 | -0.068 | 0.091 | 0.132 | -0.054 | 0.053 | -0.025 | 0.000 | 0.049 | -0.005 | 0.006 |
| block | -0.050 | -0.014 | 0.068 | 0.012 | 0.018 | -0.071 | -0.031 | 0.049 | 0.000 | 0.011 | 0.060 |
| object | -0.054 | -0.005 | -0.131 | 0.056 | -0.053 | -0.088 | 0.011 | -0.005 | 0.011 | 0.000 | 0.013 |
| coding | -0.022 | 0.021 | 0.069 | -0.067 | 0.071 | 0.158 | -0.115 | 0.006 | 0.060 | 0.013 | 0.000 |
3.2.4 Modification Indices for Model 2
Modification indices provide………………..
| lhs | op | rhs | mi | epc | sepc.lv | sepc.all | sepc.nox | |
|---|---|---|---|---|---|---|---|---|
| 34 | performance | =~ | simil | 6.763658 | 0.7945731 | 0.7945731 | 0.2502971 | 0.2502971 |
| 62 | arith | ~~ | object | 5.551414 | -0.8741210 | -0.8741210 | -0.1991496 | -0.1991496 |
| 81 | digit | ~~ | coding | 4.975650 | 1.2098255 | 1.2098255 | 0.1712424 | 0.1712424 |
| 85 | pictcomp | ~~ | coding | 4.742106 | -1.2338394 | -1.2338394 | -0.1865857 | -0.1865857 |
| 66 | simil | ~~ | pictcomp | 3.810872 | 0.9252477 | 0.9252477 | 0.1750135 | 0.1750135 |
| 52 | comp | ~~ | parang | 3.768775 | -0.8524973 | -0.8524973 | -0.1727905 | -0.1727905 |
| 73 | vocab | ~~ | parang | 3.689972 | -0.7528236 | -0.7528236 | -0.1726510 | -0.1726510 |
| 67 | simil | ~~ | parang | 3.578938 | 0.8683482 | 0.8683482 | 0.1603695 | 0.1603695 |
| 57 | arith | ~~ | vocab | 3.377455 | -0.6406313 | -0.6406313 | -0.1839884 | -0.1839884 |
| 61 | arith | ~~ | block | 3.256929 | 0.6099258 | 0.6099258 | 0.1615230 | 0.1615230 |
| 38 | info | ~~ | arith | 3.216572 | 0.6208828 | 0.6208828 | 0.1793789 | 0.1793789 |
| 70 | simil | ~~ | coding | 3.165192 | -0.9489408 | -0.9489408 | -0.1454290 | -0.1454290 |
3.2.5 Path Diagram for Model 2
The semPlot package provides a capability of drawing path diagrams for cfa and other sem models. The semPaths function will take a cfa model object and draw the diagram, with several options available. The diagram produced here takes control over font/label sizes, display of residuals, and color of paths/coefficients. These and many more control options are available. There is a challenge in producing these path diagrams to have font sizes large enough for most humans to read. I’ve taken control of the font sizes for the “edges” with a cex argument. But this causes overlap in the values if the default layout is used. I found that “circle2” worked best here.
# Note that the base plot, including standardized path coefficients plots positive coefficients green
# and negative coefficients red. Red-green colorblindness issues anyone?
# I redrew it here to choose a blue and red. But all the coefficients in this example are
# positive,so they are shown with the skyblue.
# more challenging to use colors other than red and green. not in this doc
semPaths(fit2, residuals=F,sizeMan=7,"std",
posCol=c("skyblue4", "red"),
#edge.color="skyblue4",
edge.label.cex=1.2,layout="circle2")
# or we could draw the paths in such a way to include the residuals:
#semPaths(fit1, sizeMan=7,"std",edge.color="skyblue4",edge.label.cex=1,layout="circle2")
# the base path diagram can be drawn much more simply:
#semPaths(fit2)
# or
semPaths(fit2,"std")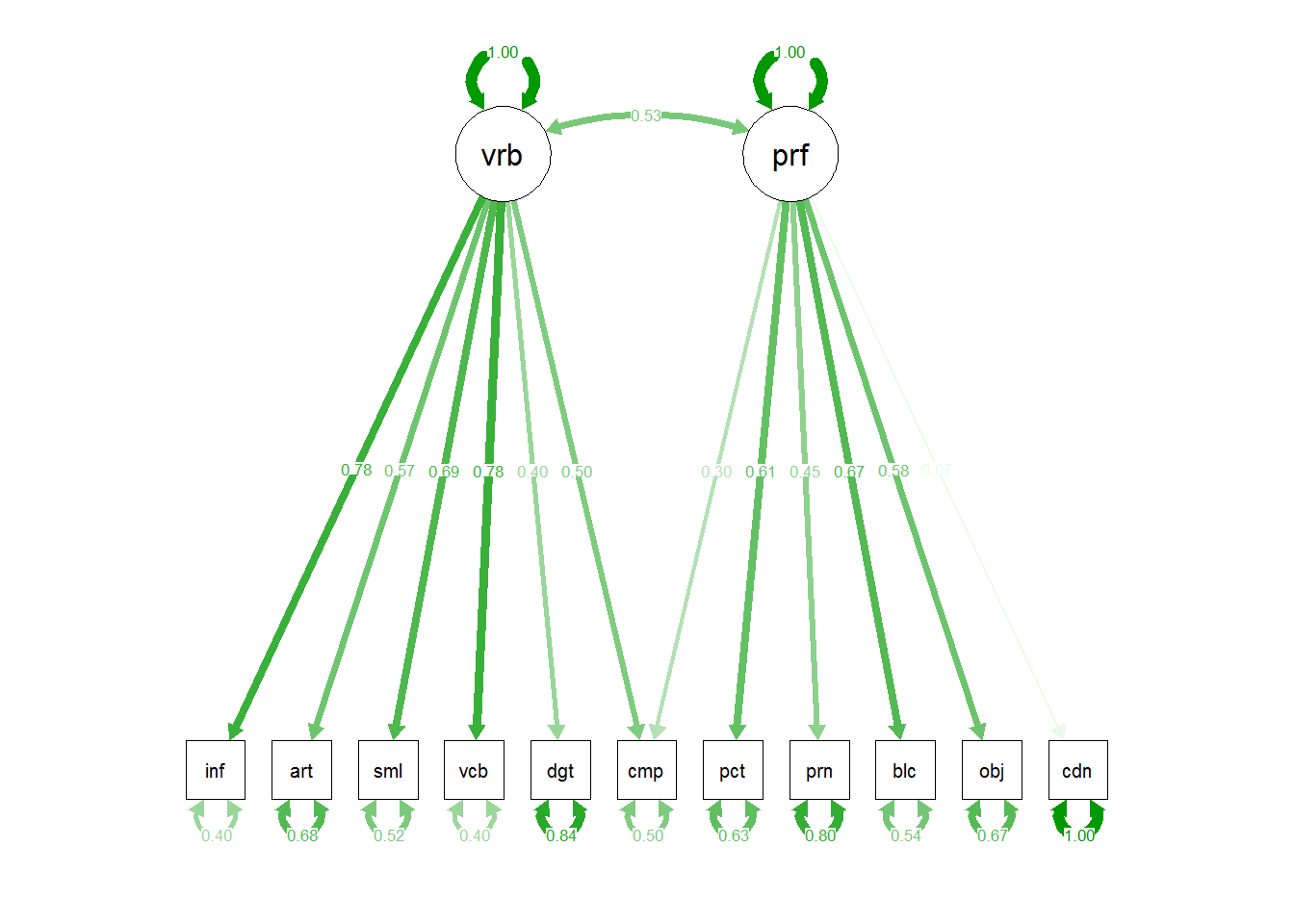
3.3 Compare Model 1 and Model 2
Model 1 is first/base. Model 2 adds a path from Perf to comp… Compare these models.
| Df | AIC | BIC | Chisq | Chisq diff | RMSEA | Df diff | Pr(>Chisq) | |
|---|---|---|---|---|---|---|---|---|
| fit2 | 42 | 9021.645 | 9097.600 | 60.64158 | NA | NA | NA | NA |
| fit1 | 43 | 9029.643 | 9102.433 | 70.63957 | 9.997993 | 0.2267534 | 1 | 0.0015671 |
3.4 An additional perspective on estimation and optimization
Subtle differences in algorithmic strategies for Structural Equation Modeling exist among software packages. Users are often familiar with only the default approaches and will only be moved to learn other approaches when the default approach fails to converge or produces problematic models. The student first confronted with these methods will often assume that the same SEM model evauated in different software (e.g., LISREL, MPlus, EQS, lavaan, sem, OpenMx) will produce the same model outcome. This may not be a safe assumption. Different “dialects” exist for the various software products. The commercial products may use algorithms that are proprietary and not available to understand their approach. The open source products (e.g., lavaan) have made source code available for inspection.
The perceptive reader will notice that the default solutions given by the three R packages employed in this document (lavaan, sem, OpenMx) all give the same values for parameter estimates and goodnes of fit statistics for the same two models run with each. However, there are slight differences in these quantities compared to the published LISREL analysis of the same data in the Tabachnik, et al textbook (-Tabachnick et al. (2019)). Other readers will have noticed that the LISREL output in this textbook matches SAS output that they may have seen with class coverage of the CFA topic. The R packages, while agreeing with each other, vary slightly from the LISREL and SAS values. The degree of difference is slight, but its existence may puzzle some students. The answer to understanding these differences goes well beyond the scope of this document and involves an advanced understanding of the computational algorithms employed. Even though all of the approaches are Maximum Liklihood methods, some optimization and estimation strategies can differ.
The lavaan package permits some insight into this with one of the argments available for the cfa function used in this chapter. The reader might want to examine the help docs for this function: ?cfa. That help page directs readers to another help document on Lavaan Options (lavoptions). There, one can find an argument that can be passed to cfa, called “mimic”. Here is the text from that section, describing the various “mimic” possibilities:
If “Mplus”, an attempt is made to mimic the Mplus program. If “EQS”, an attempt is made to mimic the EQS program. If “default”, the value is (currently) set to to “lavaan”, which is very close to “Mplus”.
The reader may have been exposed to a SAS Proc Calis approach to this problem that employed the default Method called LINEQS. The following rerun of the first model from this chapter above, employs the mimic argument to be specified as “EQS”. The product of this model is a set of parameter values and fit statistics (e.g., Chi Sq) that match the SAS output (and the Tabachnick et al LISREL output) exactly. Demonstrating the equivalence with the addition of the mimic argument does not fully explain why such differences originally existed, but that, again, is well beyond the scope of this document. The reference section to this document includes some articles that address these differences (El-Sheikh et al., 2017; Narayanan, 2012). Rosseel (2012) has discussed use of the ‘mimic’ function that guides lavaan to emulate the approach of some of the commercial products. His website is also a valuable resource on this [http://lavaan.ugent.be/].
wisc.model1 <- "verbal =~ info + comp + arith + simil + vocab + digit
performance =~ pictcomp + parang + block + object + coding"fit1eqs <- lavaan::cfa(wisc.model1, data=wisc2,std.lv=TRUE, mimic="EQS")
summary(fit1eqs, fit.measures=T,standardized=T)## lavaan 0.6-19 ended normally after 25 iterations
##
## Estimator ML
## Optimization method NLMINB
## Number of model parameters 23
##
## Number of observations 175
##
## Model Test User Model:
##
## Test statistic 70.236
## Degrees of freedom 43
## P-value (Chi-square) 0.005
##
## Model Test Baseline Model:
##
## Test statistic 516.237
## Degrees of freedom 55
## P-value 0.000
##
## User Model versus Baseline Model:
##
## Comparative Fit Index (CFI) 0.941
## Tucker-Lewis Index (TLI) 0.924
##
## Loglikelihood and Information Criteria:
##
## Loglikelihood user model (H0) -4497.337
## Loglikelihood unrestricted model (H1) -4462.018
##
## Akaike (AIC) 9040.675
## Bayesian (BIC) 9113.465
## Sample-size adjusted Bayesian (SABIC) 9040.631
##
## Root Mean Square Error of Approximation:
##
## RMSEA 0.060
## 90 Percent confidence interval - lower 0.033
## 90 Percent confidence interval - upper 0.085
## P-value H_0: RMSEA <= 0.050 0.239
## P-value H_0: RMSEA >= 0.080 0.101
##
## Standardized Root Mean Square Residual:
##
## SRMR 0.059
##
## Parameter Estimates:
##
## Standard errors Standard
## Information Expected
## Information saturated (h1) model Structured
##
## Latent Variables:
## Estimate Std.Err z-value P(>|z|) Std.lv Std.all
## verbal =~
## info 2.212 0.201 10.997 0.000 2.212 0.760
## comp 2.048 0.212 9.682 0.000 2.048 0.691
## arith 1.304 0.173 7.534 0.000 1.304 0.565
## simil 2.238 0.226 9.911 0.000 2.238 0.703
## vocab 2.257 0.202 11.193 0.000 2.257 0.770
## digit 1.056 0.213 4.952 0.000 1.056 0.390
## performance =~
## pictcomp 1.747 0.244 7.166 0.000 1.747 0.595
## parang 1.257 0.226 5.566 0.000 1.257 0.473
## block 1.851 0.223 8.287 0.000 1.851 0.683
## object 1.609 0.237 6.780 0.000 1.609 0.566
## coding 0.208 0.256 0.811 0.417 0.208 0.072
##
## Covariances:
## Estimate Std.Err z-value P(>|z|) Std.lv Std.all
## verbal ~~
## performance 0.589 0.076 7.792 0.000 0.589 0.589
##
## Variances:
## Estimate Std.Err z-value P(>|z|) Std.lv Std.all
## .info 3.586 0.511 7.014 0.000 3.586 0.423
## .comp 4.599 0.590 7.793 0.000 4.599 0.523
## .arith 3.623 0.424 8.547 0.000 3.623 0.681
## .simil 5.125 0.667 7.680 0.000 5.125 0.506
## .vocab 3.507 0.511 6.866 0.000 3.507 0.408
## .digit 6.198 0.686 9.030 0.000 6.198 0.848
## .pictcomp 5.558 0.764 7.276 0.000 5.558 0.646
## .parang 5.494 0.664 8.275 0.000 5.494 0.777
## .block 3.916 0.646 6.066 0.000 3.916 0.533
## .object 5.499 0.726 7.578 0.000 5.499 0.680
## .coding 8.206 0.882 9.309 0.000 8.206 0.995
## verbal 1.000 1.000 1.000
## performance 1.000 1.000 1.000